Linear Support Vector Machine¶
This material is heavily based on the popular Standford CS231n lecture material. Please check on their website for more detailed information.
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams.update({
"text.usetex": True,
"font.family": "sans-serif",
"font.size": 10,
})
from utils import *
np.set_printoptions(precision=5, suppress=True)
Class SVM¶
This is the Python class implementation of the linear SVM:
There two metods:
train(X, Y)andY = predict(X).Matrix
self.Walso includes the bias vector (bias tricks).Training is done by using stochastic gradient descent method or gradient descent method.
Let us define the score function as linear combination of all feature values.
\( F = X W \)
\(L_i = \sum_{j \neq Y_i} \max(0, F_{i,j}-F_{i,Y_i} + 1)\)
\( L=\underbrace{\frac{1}{N} \sum_{i=1}^N L_{i}}_{\text {data loss }}+\underbrace{\lambda R(W)}_{\text {regularization loss }} \)
\( R(W) = \sum_{k=1}^C \sum_{l=1}^{D+1} W_{l, k}^{2} \)
where:
the dataset contains \(N\) samples of data
each sample has \(D\) features (dimensionalities) and \(C\) labels/classes (distinct categories)
\(X \in \mathbb{R}^{N \times (D+1)}\) contains the features from N samples of data
\(X_i \in \mathbb{R}^{D+1}\) contains the features of the \(i\)-th sample (\(i\)-th row of \(X\))
\(Y \in \mathbb{R}^{N}\) contains the correct classes from N samples of data
\(Y_i \in \mathbb{R}\) contains the correct class of the \(i\)-th sample (\(i\)-th row of \(Y\))
\(F_{i,j} \in \mathbb{R}\) is the score for \(i\)-th sample and \(j\)-th class (\(i\)-th row and \(j\)-th column)
\(F_{i,Y_i} \in \mathbb{R}\) is the score for the correct class of \(i\)-th sample
\(L \in \mathbb{R}\) is the total loss
\(L_i \in \mathbb{R}\) is the loss of \(i\)-th sample
\(W \in \mathbb{R}^{(D+1) \times C}\) is the weight matrix (with augmented bias vector in the last row)
\(R \in \mathbb{R}\) is the regularization loss
Note: the words “label” and “class” have similar meaning!
from numba import njit, prange
class SVM():
@staticmethod
@njit(parallel=True, fastmath=True)
def svm_loss(W, X, Y, reg):
"""
Structured SVM loss function, naive implementation (with loops).
Inputs have dimension D, there are C classes, and we operate on minibatches
of N examples.
Inputs:
- W: A numpy array of shape (D+1, C) containing weights.
- X: A numpy array of shape (N, D) containing a minibatch of data.
- Y: A numpy array of shape (N,) containing training labels; y[i] = c means
that X[i] has label c, where 0 <= c < C.
- reg: (float) regularization strength
Returns a tuple of:
- loss: loss as single float
- dW: gradient with respect to weights W; an array of same shape as W
References:
- https://github.com/lightaime/cs231n
- https://github.com/mantasu/cs231n
- https://github.com/jariasf/CS231n
"""
X = np.hstack((X, np.ones((X.shape[0],1)))) # the last column is 1: to allow augmentation of bias vector into W
dW = np.zeros(W.shape) # initialize the gradient as zero
# compute the loss and the gradient
C = W.shape[1]
N = X.shape[0]
loss = 0.0
F = X@W
for i in prange(N):
Fi = F[i]
Fyi = Fi[Y[i]]
Li = Fi - Fyi + 1 #
Li[np.where(Li < 0)] = 0 # max(0, ...)
loss += np.sum(np.delete(Li, Y[i]))
for j in prange(C):
if Li[j] > 0 and j != Y[i]:
dW[:, j] += X[i] # update gradient for incorrect label
dW[:, Y[i]] -= X[i] # update gradient for correct label
loss = loss / N + reg * np.sum(W * W) # add regularization to the loss.
dW = dW / N + 2 * reg * W # append partial derivative of regularization term
return loss, dW
@staticmethod
def svm_loss_vectorized(W, X, Y, reg):
"""
Structured SVM loss function, naive implementation (with loops).
Inputs have dimension D, there are C classes, and we operate on minibatches
of N examples.
Inputs:
- W: A numpy array of shape (D+1, C) containing weights.
- X: A numpy array of shape (N, D) containing a minibatch of data.
- Y: A numpy array of shape (N,) containing training labels; y[i] = c means
that X[i] has label c, where 0 <= c < C.
- reg: (float) regularization strength
Returns a tuple of:
- loss: loss as single float
- dW: gradient with respect to weights W; an array of same shape as W
References:
- https://github.com/lightaime/cs231n
- https://github.com/mantasu/cs231n
- https://github.com/jariasf/CS231n
"""
X = np.hstack((X, np.ones((X.shape[0],1)))) # the last column is 1: to allow augmentation of bias vector into W
N = X.shape[0]
# scores
F = X @ W
# Scores for correct labels
Fy = F[range(N), Y].reshape(-1,1)
# compute the loss
L = F - Fy + 1
L = np.maximum(0, L) # max(0, ...)
L_ = L.copy() # keep the original
L[range(N) , Y] = 0 # exclude the correct labels
loss = np.sum(L) / N + reg * np.sum(W * W)
# compute the gradient
dW = (L_ > 0).astype(float) # positive for incorrect labels
dW[range(N), Y] = dW[range(N), Y] - dW.sum(axis=1) # negative for correct labels
dW = X.T @ dW / N + 2 * reg * W # gradient with respect to W
return (loss, dW)
def train(self, X, Y, learning_rate=1e-3, reg=1e-5, num_iters=100, batch_size=200, verbose=True, verbose_step=1000):
'''
Train this linear classifier using stochastic gradient descent.
Setting the 'batch_size=None" turns of the stochastic feature.
Inputs:
- X: A numpy array of shape (N, D) containing training data; there are N
training samples each of dimension D.
- y: A numpy array of shape (N,) containing training labels; y[i] = c
means that X[i] has label 0 <= c < C for C classes.
- learning_rate: (float) learning rate for optimization.
- reg: (float) regularization strength.
- num_iters: (integer) number of steps to take when optimizing
- batch_size: (integer) number of training examples to use at each step.
- verbose: (boolean) If true, print progress during optimization.
- verbose_steps: (integer) print proress once every verbose_steps
Outputs:
A list containing the value of the loss function at each training iteration.
'''
N, D = X.shape
C = len(np.unique(Y))
# lazily initialize W
self.W = 0.001 * np.random.randn(D + 1, C) # dim+1, to bias vector is augmented into W
# Run stochastic gradient descent to optimize W
loss_history = []
for it in range(num_iters):
X_batch = None
y_batch = None
# Sample batch_size elements from the training data and their
# corresponding labels to use in this round of gradient descent.
# Store the data in X_batch and their corresponding labels in
# y_batch; after sampling X_batch should have shape (dim, batch_size)
# and y_batch should have shape (batch_size,)
if batch_size is not None:
batch_indices = np.random.choice(N, batch_size, replace=False)
X_batch = X[batch_indices]
y_batch = Y[batch_indices]
loss, grad = self.svm_loss_vectorized(self.W, X_batch, y_batch, reg)
else:
loss, grad = self.svm_loss_vectorized(self.W, X, Y, reg)
loss_history.append(loss)
# Update the weights using the gradient and the learning rate.
self.W = self.W - learning_rate * grad
if verbose and it % verbose_step == 0 or it == num_iters - 1:
print('iteration %d / %d: loss %f' % (it, num_iters, loss), flush=True)
return loss_history
def predict(self, X):
"""
Use the trained weights of this linear classifier to predict labels for
data points.
Inputs:
- X: A numpy array of shape (N, D) containing training data; there are N
training samples each of dimension D.
Returns:
- Y_pred: Predicted labels for the data in X. y_pred is a 1-dimensional
array of length N, and each element is an integer giving the predicted
class.
"""
X = np.hstack((X, np.ones((X.shape[0],1))))
Y_pred = np.zeros(X.shape[0])
scores = X @ self.W
Y_pred = scores.argmax(axis=1)
return Y_pred
Breast Cancer Wisconsin¶
This dataset is publicly available and can be downloaded from this link.
This dataset is also available as one of scikit-learn example datasets.
The original labels are characters: M and B. For the SVM to work, the labels must be numbers. Hence, we change the labels:
Mis replaced with1Bis replaced with0
Load the Dataset¶
data = np.loadtxt("./datasets/breast_cancer/wdbc.data", delimiter=",", dtype=str)
X = np.float32(data[:, 2:12]) # 10 dimensions
# Diagnosis (M = malignant, B = benign)
Y = np.zeros(X.shape[0], dtype=np.int32)
Y[np.where(data[:,1]=='M')] = 1
Y[np.where(data[:,1]=='B')] = 0
print("Dimension numbers :", X.shape[1])
print("Number of data :", X.shape[0])
print("Labels :", np.unique(Y))
Dimension numbers : 10
Number of data : 569
Labels : [0 1]
Split The Dataset for Training and Test¶
X_train = X[0:400, :]
y_train = Y[0:400]
X_test = X[401:, :]
Y_test = Y[401:]
num_test = X_test.shape[0]
Train the Classifier¶
classifier = SVM()
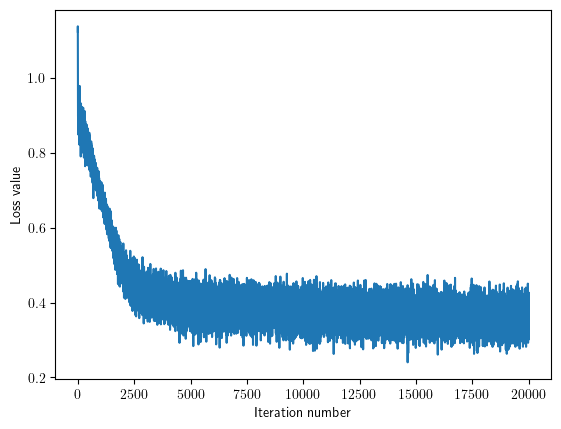
loss_hist = classifier.train(X_train, y_train, learning_rate=1e-6,batch_size=200, reg=1, num_iters=20000, verbose_step=2000)
iteration 0 / 20000: loss 1.122593
iteration 2000 / 20000: loss 0.501769
iteration 4000 / 20000: loss 0.433979
iteration 6000 / 20000: loss 0.365286
iteration 8000 / 20000: loss 0.363138
iteration 10000 / 20000: loss 0.392463
iteration 12000 / 20000: loss 0.349064
iteration 14000 / 20000: loss 0.355593
iteration 16000 / 20000: loss 0.414111
iteration 18000 / 20000: loss 0.355115
iteration 19999 / 20000: loss 0.328130
Plot the Loss¶
Plot the loss as a function of iteration number.
plt.plot(loss_hist)
plt.xlabel('Iteration number')
plt.ylabel('Loss value')
plt.show()
Test the Classifier¶
Evaluate the performance on both the training and validation set.
Y_train_pred = classifier.predict(X_train)
print('training accuracy: %f' % (np.mean(y_train == Y_train_pred), ))
Y_test_pred = classifier.predict(X_test)
print('validation accuracy: %f' % (np.mean(Y_test == Y_test_pred), ))
training accuracy: 0.857500
validation accuracy: 0.892857
Handwritten Digits¶
This dataset is publicly available and can be downloaded from this link.
Training Data¶
data = np.loadtxt("./datasets/handwritten_digits/optdigits.tra", delimiter=",", dtype=int)
X_train = np.int32(data[:, 0:-1])
y_train = np.int32(data[:, -1])
print("Dimension numbers :", X_train.shape[1])
print("Number of data :", X_train.shape[0])
print("Labels :", np.unique(y_train))
Dimension numbers : 64
Number of data : 3823
Labels : [0 1 2 3 4 5 6 7 8 9]
Test Data¶
for i in range(100):
X_train_ = X_train[i,:].reshape(8, 8)
X_train_ = np.abs(255.0 - 255.0 / 16.0 * X_train_)
plt.subplot(20, 10, i + 1)
# Rescale the weights to be between 0 and 255
plt.imshow(X_train_.astype('uint8'), cmap='Greys')
plt.axis('off')
data = np.loadtxt("./datasets/handwritten_digits/optdigits.tes", delimiter=",", dtype=int)
X_test = np.int32(data[:, 0:-1])
y_test = np.int32(data[:, -1])
print("Dimension numbers :", X_test.shape[1])
print("Number of data :", X_test.shape[0])
print("Labels :", np.unique(y_test))
Dimension numbers : 64
Number of data : 1797
Labels : [0 1 2 3 4 5 6 7 8 9]
Train the Classifier¶
classifier = SVM()
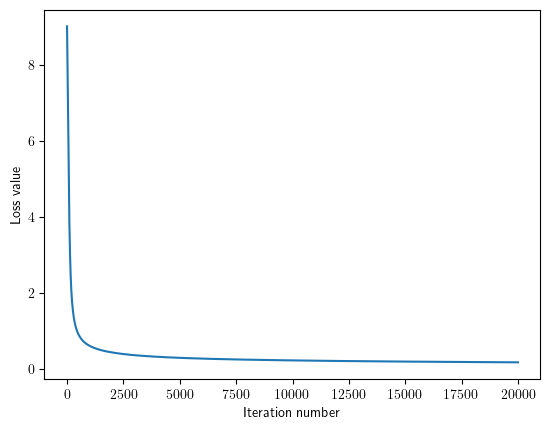
loss_hist = classifier.train(X_train, y_train, learning_rate=1e-5, batch_size=None, num_iters=20000, verbose_step=5000)
iteration 0 / 20000: loss 9.024547
iteration 5000 / 20000: loss 0.289658
iteration 10000 / 20000: loss 0.222047
iteration 15000 / 20000: loss 0.191010
iteration 19999 / 20000: loss 0.171675
Plot the Loss¶
Plot the loss as a function of iteration number.
plt.plot(loss_hist)
plt.xlabel('Iteration number')
plt.ylabel('Loss value')
plt.show()
Test the Classifier¶
Evaluate the performance on both the training and validation set.
y_train_pred = classifier.predict(X_train)
print('training accuracy: %f' % (np.mean(y_train == y_train_pred), ))
y_test_pred = classifier.predict(X_test)
print('validation accuracy: %f' % (np.mean(y_test == y_test_pred), ))
training accuracy: 0.966780
validation accuracy: 0.933779
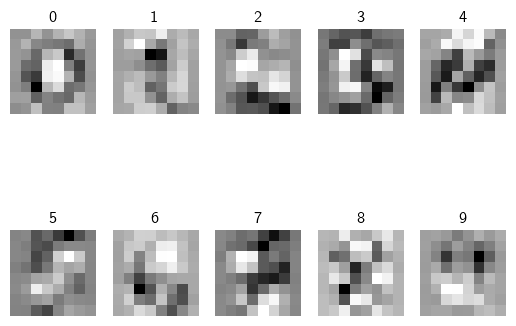
Visualize the Learned Weights¶
Take matrix
Wand strip out the bias.For each class, reshape the matrix back into 2D arrays
Plot the 2D arrays (matrices) as images.
w = classifier.W[:-1, :] # strip out the bias
w = w.reshape(8, 8, 10)
w_min, w_max = np.min(w), np.max(w)
classes = ['0', '1', '2', '3', '4', '5', '6', '7', '8', '9']
for i in range(10):
plt.subplot(2, 5, i + 1)
# Rescale the weights to be between 0 and 255
wimg = 255.0 * (w[:, :, i].squeeze() - w_min) / (w_max - w_min)
plt.imshow(wimg.astype('uint8'), cmap='Greys')
plt.axis('off')
plt.title(classes[i])